This blog post adds to an earlier series that discusses the Genome Archive (GenArk) assembly hubs. To help users both to find Genome Archive (GenArk) hubs and to inform us what GCA/GCF accessions to add to the collection, we have created a new genome assembly request page: http://genome.ucsc.edu/assemblyRequest.html
Last year we announced the creation of a new collection of GenArk assembly hubs. GenArk hubs are constructed from NCBI Genbank GCA/GCF accessioned assembly data, for instance where GCF_001984765.1 is the accession for an American beaver assembly. When present in UCSC’s GenArk collection, these genome browsers can be loaded instantly with direct links (e.g., http://genome.ucsc.edu/h/GCF_001984765.1), and they come ready with dynamically invoked BLAT and PCR servers, enabling searching for sequences and primers. The first released GenArk hubs were organized into phylogenetic groups, for example, all bird assemblies were listed here.
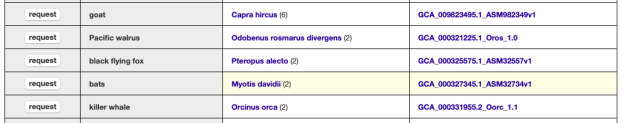
Our newly added genome assembly request page displays which assemblies are available for viewing, and presents a single-click “request” button to send an email to UCSC to add any GCA/GCF assembly available at NCBI not yet part of the GenArk collection.
GenArk currently has over 1,700 assembly hubs available for browsing at the click of a “view” button. To view only those in the current collection, you can use the middle “select assembly type to display” option and remove the “Request browser” checkbox, and only completed browsers will show. Upon first visiting the page shows only the first 500 assemblies. Use the “select assembly type to display” to “show all” assemblies if you do not see your assembly. Click “view” to launch a specific genome browser listed on the page. This earliest version of the page has some performance issues, and selections may take some time, a future improvement planned is to present an active wait cursor when the page is busy filtering results.
By using the “choose clade to view/hide” option it is possible to subselect groups, such as only displaying plants, when narrowing down which genomes may exist or could be requested.
The first “show/hide columns” option on the page enables displaying additional metadata, such as adding the assembly build date or Biosample number, where links exist back to NCBI for more information.
If NCBI does not have a GCA/GCF accession for your desired assembly then our scripts will not be able to pull the data and generate the GenArk addition. Such new assemblies will need to be submitted to NCBI first, after which you can then notify UCSC. You can find directions at NCBI for how to submit new genomes: https://www.ncbi.nlm.nih.gov/assembly/docs/submission/ Also, please review the UCSC GenArk Blog posts for more information on accessing and using tools on GenArk hubs.
If after reading this blog post you have any public questions, please email genome@soe.ucsc.edu. All messages sent to that address are archived on a publicly accessible forum. If your question includes sensitive data, you may send it instead to genome-www@soe.ucsc.edu.