The previous parts of this series (part 1 and part 2) focused on how to use the Genome Browser to obtain data, and for this third and final post we’re gonna divert from that theme and talk about how to control the track image itself. Note: We now have an API which can also perform many of these functions.
Standard procedure for obtaining images of the browser is to configure the view exactly as you want, and then use the “View->PDF/PS” option in the menu bar in order to download a PDF or PostScript of your image. In addition to this method, you can generate PNG images on the fly with the following hgRenderTracks template:
http://genome.ucsc.edu/cgi-bin/hgRenderTracks?parameters
Parameters should be replaced by the URL key-value pairs that the main track display, hgTracks, understands, like ‘db=hg19’ or ‘knownGene=pack’. For example, to compare the transcripts provided by NCBI to UCSC’s own alignments of the transcripts at the ABO locus, you can use the following URL and the cURL program to download a PNG file:
curl 'http://genome.ucsc.edu/cgi-bin/hgRenderTracks?db=hg19&position=chr9:136130563-136150630&hideTracks=1&refSeqComposite=pack&ncbiRefSeqCurated_sel=1&ucscRefSeqView=pack&refGene_sel=1&pubs=pack' > example.png
Opening example.png in your favorite image viewer will display the following image:

There are many additional parameters described on the Sharing your custom track section of the custom tracks help page. Using these parameters you can configure hgRenderTracks to display any combinations of tracks, and along the hgt.customText parameter, also show your custom tracks with them.
To illustrate, if I have the following custom track:
browser hide all browser gold=pack browser gap=pack browser visibility=pack chr1 1000 2000 chr1 2100 3000 chr1 3100 4000 …
Hosted on the web at http://genome-test.soe.ucsc.edu/~chmalee/exCustomTrack.bed, then I can tell hgRenderTracks to load this file with the hgt.customText parameter like so:
http://genome.ucsc.edu/cgi-bin/hgRenderTracks?db=hg38&position=chr1:1-100000&hgt.customText=http://genome-test.soe.ucsc.edu/~chmalee/exCustomTrack.bed

The “browser” lines at the beginning of the custom track indicate which native tracks to turn on along their visibilities, while the “hide all” line turns all the other native tracks off. In addition to these basic instructions there are many more examples on the UCSC Genome Browser Wiki.
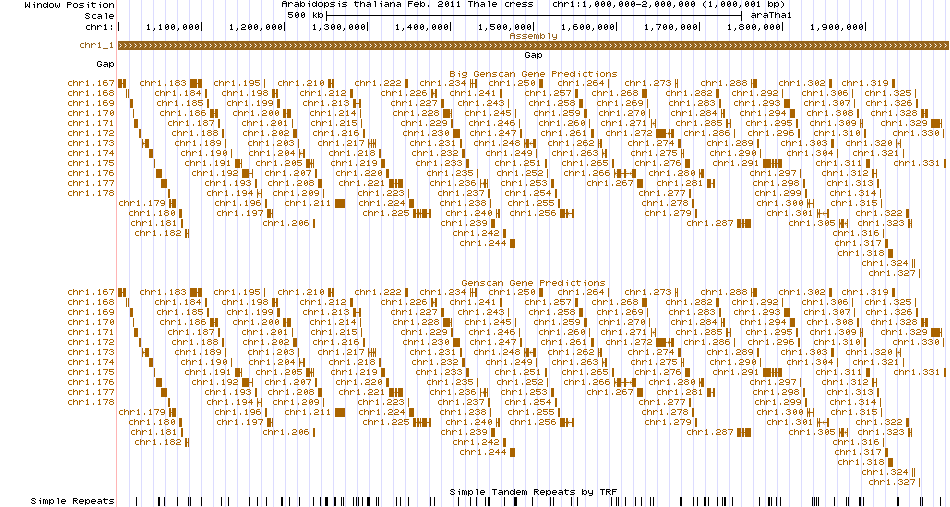
What about when you want to view a genome and annotations not hosted on our site? If you have a FASTA file of your genome available, you can use faToTwoBit to convert your genome into a 2bit file, then make an assembly hub out of your data. Once you’ve created your hub, you can view the hub with the hubUrl setting. As an example, I have hosted an assembly hub for Arabadopsis thaliana here, and I can view the hub via a single URL like so:
https://genome.ucsc.edu/cgi-bin/hgTracks?genome=araTha1&hubUrl=https://genome-test.gi.ucsc.edu/~chmalee/araTha1/plantAraTha1/hub.txt.
If your data needs to stay behind your local firewall, then you can use the GBiB and GBiC products so you can set up your own “copy” of the UCSC Genome Browser that meets your privacy needs.
Further Reading:
- UCSC Genome Browser Downloads server
- GenomeWiki
- Track Image Parameters
- Custom Tracks
- Track Hubs
- Viewing private data on the Genome Browser
If after reading this blog post you have any public questions, please email genome@soe.ucsc.edu. All messages sent to that address are archived on a publicly accessible forum. If your question includes sensitive data, you may send it instead to genome-www@soe.ucsc.edu.
