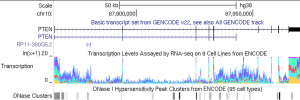
Genome Browser screen shot of the GRCh38 (hg38) human assembly showing the GENCODE Basic track opened in the PTEN region on chromosome 10.
As of Monday, July 29, 2015, the UCSC Genome Browser will use the GENCODE v22 comprehensive gene set as its default gene set on the human genome assembly GRCh38 (hg38), replacing the previous default set of genes created here at UCSC using code written by Jim Kent. This track, which is labeled as “GENCODE Basic” in the Genes and Gene Predictions track group, replaces UCSC Genes track as the default gene set. We’re making this change in recognition of the value of reducing the number of competing gene sets used by the bioinformatics community. With this change we will be using the same set of genes as Ensembl, reducing the potential for confusion, especially in clinical settings.
We’ve kept the same familiar UCSC Genes schema for the new gene set, using nearly all the same table names and fields that appeared in earlier versions of UCSC Genes. Hopefully this will make the transition to the new GENCODE models easier. Every transcript in the new set has both a UCSC ID and a GENCODE transcript ID. There are a couple of new tables: knownCds, which has the coding frame numbers for each gene, and knownToMrna, which captures the association to GenBank mRNAs. A couple tables are no longer present: knownGeneTxMrna and knownGeneTxPep.
By default, we display only the transcripts tagged as “basic” by the GENCODE Consortium. However, all the transcripts in the GENCODE comprehensive set are present in the tables. You can view them in the browser by selecting “show comprehensive set” in the “Show” section of the track’s description page. On that same page, you can also configure the browser to label the genes with the GENCODE transcript IDs by selecting “GENCODE Transcript ID” label option.
The new gene set has 195,178 total transcripts, compared with 104,178 in the previous UCSC Genes version. The total number of canonical genes, now defined using the GENCODE gene loci ( ENSG* identifiers), has increased from 48,424 to 49,534.
Comparing the previous gene set with the new version:
- 9,459 transcripts are identical.
- 22,088 transcripts were not carried forward to the new version.
- 43,681 have consistent splicing, but changes in the UTR.
- 28,950 transcripts overlap with those in the previous set, but have
at least one different splice.
We plan to continue using the previous UCSC computational pipeline to generate the default gene set on the mouse assembly, GRCm38 (mm10), for the foreseeable future. We will also periodically update the old UCSC-computed gene set on the human GRCh38 assembly as an ancillary track (“Old UCSC Genes”) without the rich set of link-outs we maintain for the default gene set.
If after reading this blog post you have any public questions, please email genome@soe.ucsc.edu. All messages sent to that address are archived on a publicly accessible forum. If your question includes sensitive data, you may send it instead to genome-www@soe.ucsc.edu.
